 An oft-repeated message from scientists involved with the Large Hadron Collider (LHC), mapping the human genome, the search for extraterrestrial life and other vast scientific projects, such as supercomputing experiments is that the tera-bytes, peta-bytes, perhaps even the yotta-bytes of data generated by large-scale projects is hard to handle, to say the least.
An oft-repeated message from scientists involved with the Large Hadron Collider (LHC), mapping the human genome, the search for extraterrestrial life and other vast scientific projects, such as supercomputing experiments is that the tera-bytes, peta-bytes, perhaps even the yotta-bytes of data generated by large-scale projects is hard to handle, to say the least.
Not only has there to be a way to manage the outpouring of data from the sensors, monitors, and arrays, but this data has to be channelled through the appropriate digital conduits, dumped into robust databases, processed in parallel, and finally interpreted by the scientists themselves. Only then can DNA be fully unravelled, ET phoned home about, and god in the particles revealed…
A team at the Federal University of Rio de Janeiro and the Fluminense Federal University in Brazil hope to one day help big science address this big problem by finding ways to manage the scientific workflow on large-scale experiments.
Computer scientist Marta Mattoso and colleagues explain that, ‘One of the main challenges of scientific experiments is to allow scientists to manage and exchange their scientific computational resources (data, programs, models, etc.).’ They add that effective management of such experiments requires experiment specification techniques, workflow derivation heuristics and provenance mechanisms. In other words, there has to be a way to characterise an experiment’s life cycle into three phases: composition, execution, and analysis. To be frank, the same scientific ethics apply to all experiments from the simplest kitchen science fair project to the LHC and beyond by way of a million laboratory benches.
However, while most scientists and students are familiar with this format of introduction, method, conclusion, the researchers point out that research into improving efficiency and efficacy in big science projects has largely focused on execution and analysis, the method and conclusion, rather than considering the scientific experiment as a whole. For example, Mattoso and colleagues point out that the myGrid project back in the early 2000s was one of the first works to define a scientific experiment life cycle and I worked with the myGrid team on information materials at the time. Their approach used six phases: construction, discovery, personalisation, execution and management. However, they stick to the execution and analysis aspects of the project. Others have focused on large-scale projects in a similarly limited way.
Mattoso and colleagues have proposed an approach for managing large-scale experiments based on provenance gathering during all phases of the life cycle. ‘We foresee that such approach may aid scientists to have more control on the trials of the scientific experiment,’ they say.
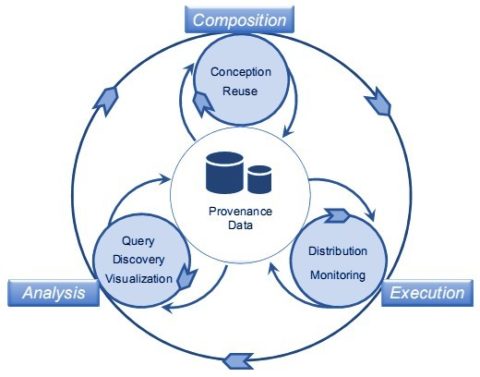
In their model, each phase – composition, execution and analysis – has an independent cycle within the lifecycle as a whole, wheels within wheels, one might say. Each independent cycle runs at distinct points in the experiment and is associated with its own explicit meta data.

‘The composition phase is responsible for structuring and setting up the whole experiment, establishing the logical sequence of activities, the type of input data and parameters that should be provided, and the type of output data that are generated,’ the team explains. This cycle has within it the conception sub-phase responsible for setting up the experiment and the reuse phase responsible for retrieving an existing protocol and adapting it to new ends. The execution phase is responsible for making the experiment happen and represents a solid workflow instance and within this cycle there are two sub-phases: distribution and monitoring. The third cycle within the whole is the analysis phase, which is responsible for studying the data generated. This phase might be carried out by query or by visualisation.
The ultimate output of this wheels-within-wheels lifecycle is one of two possibilities: (a) the result upholds the scientific hypothesis (b) the hypothesis is refuted by the results.
In both cases scientists will then need a new workflow lifecycle to either validate the results or devise a new hypothesis that takes into account the new findings. The former is the stuff of peer review and publication, the latter hopefully posits that most intriguing of scientific exclamations: ‘That’s funny!?‘
The raison d’etre for the Open Notebook Science movement is brought to mind by the team’s concluding remarks about reproducibility and validity encapsulated by a scientific lifecycle that lays bare the experimental provenance and entire history:
‘The authors believe that by supporting provenance throughout all phases of the experiment life cycle, the provenance data will help scientists not only to analyse results to verify if their hypothesis can be confirmed or refuted, but also may assist them to protect the integrity, confidentiality, and availability of the relevant and fundamental information to reproduce the large scale scientific experiment and share its results (i.e., the complete history of the experiment).’
![]() Mattoso, M., Werner, C., Travassos, G., Braganholo, V., Ogasawara, E., Oliveira, D., Cruz, S., Martinho, W., & Murta, L. (2010). Towards supporting the life cycle of large scale scientific experiments International Journal of Business Process Integration and Management, 5 (1) DOI: 10.1504/IJBPIM.2010.033176
Mattoso, M., Werner, C., Travassos, G., Braganholo, V., Ogasawara, E., Oliveira, D., Cruz, S., Martinho, W., & Murta, L. (2010). Towards supporting the life cycle of large scale scientific experiments International Journal of Business Process Integration and Management, 5 (1) DOI: 10.1504/IJBPIM.2010.033176