
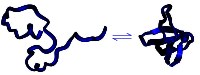 Are films ferroelectric?
Are films ferroelectric?
Discipline for gold nanocrystals
Photosynthetic system
Dissecting the atom
Catalytic clues
SAXS and the water channel
Digging in the dirt
X-ray movies
Protein based sensors
A new class of sensor for important biomolecules, pollutants, and compounds
relevant to defence is on the horizon thanks to studies into how proteins
fold into their active shape.
Kevin Plaxco of the University of California Santa Barbara and colleagues
used BioCAT and BESSRC/XOR beamlines at the Advanced Photon Source and
beamline 4-2 at the Stanford Synchrotron Radiation Laboratory to probe the
nature of the random coiling of proteins and to compare the dimensions of
these coils with chemically unfolded proteins. Their research offers
important clues about the behavior of proteins and will help their effort to
exploit proteins in real-time optical and electronic sensors for the
biotechnology industry and in biomedicine.
Previous research using various spectroscopic techniques, such as nuclear
magnetic resonance (NMR) spectroscopy, led to the tantalizing conclusion
that many proteins can keep their shape even in chemical conditions that
would quickly denature, or break down, the structure of lesser proteins. For
instance, NMR showed that even in urea or guanidine hydrochloride, two
strongly denaturing chemical reagents, several proteins display ordered
structure. These findings suggested that this residual structure might play
a role in the initial folding kinetics and thermodynamics of certain
proteins.
However, small angle x-ray scattering studies appear to give a different
picture of these proteins, revealing them to behave in certain respects as
random coils completely devoid of any organized structure.
Plaxco and his colleagues hoped to explain this discrepancy using a detailed
study of several proteins that researchers claimed were robust to
denaturation. He explains that if there were residual structure, as the
spectroscopic studies suggest, then the dimensions of these tough proteins
would not follow the same rules as a randomly coiled peptide chain. "A
hallmark of random-coil behavior is a power-law relationship between peptide
chain length and the ensemble average radius of gyration (RG)," There is a
highly characteristic relationship between the length of a polymer, and the
dimension of the bundle it forms. This power-law relationship can
distinguish between a random coil and a coil with structure. Plaxco suggests
a simple analogy. Take the mean distance between the two ends of a piece of
yarn floating in a swimming pool, he suggests, they will naturally increase
for longer pieces of yarn. But, if the yarn is wrapped into a tight ball,
the end-to-end distance will be related to the length of the yarn by its
"1/3 power". If, on the other hand, the yarn is unwound and adopts an
expanded, random structure, the increase in end-to-end distance will follow
the "0.588 power" of the length instead. This 0.588 power rule holds for any
fully random polymer, whether a length of yarn or a sub-microscopic protein
molecule.
Plaxco and his colleagues investigated 28 proteins that had a claim to
denaturation resilience and found that only two deviated from the expected
0.588 behavior of a random coil. If there were true resilience, he explains,
one would expect all of the proteins to deviate significantly from this
relationship.
"It appears that, if there is residual structure in the unfolded states -
structure that might guide the folding process - it is too subtle to affect
the mean overall dimensions of the unfolded state," says Plaxco, "This
should put strong constraints on our mental model of the unfolded state and,
we think, should have profound consequences for our understanding of
folding."
Plaxco suggests that a clearer understanding of protein folding could
ultimately allow him and others to develop new simple and efficient sensors
based on the folding of proteins, peptides and DNA when they come into
contact with compounds of interest in a sample. Indeed, the motivation for
this research was Plaxco's efforts to build optical sensors based on the
binding-induced folding of normally unfolded proteins. "If the unfolded
state is truly random, then this binding-induced folding represents the
largest possible change in protein conformation (from random to perfectly
structured), suggesting that they are an ideal means of coupling binding
events with optical or electronic outputs" he explains.
Sensors based on this behavior would be inexpensive and easy to use and so
might allow sensor technology to become more readily accessible in home
testing kits and in the sensors for the developing world.
See Jonathan E. Kohn,1 Ian S. Millett,2 Jaby Jacob,3,4 Bojan Zagrovic,5
Thomas M. Dillon,6 Nikolina Cingel,6 Robin S. Dothager,3 Soenke Seifert,7 P.
Thiyagarajan,4 Tobin R. Sosnick,3,8 M. Zahid Hasan,9 Vijay S. Pande,5 Ingo
Ruczinski,10 Sebastian Doniach,2 and Kevin W. Plaxco,1,6,5 "Random-coil
behavior and the dimensions of chemically unfolded proteins," Proc Natl Acad
Sci 101, 12491-12496 (2004).
Author affiliation 1Interdepartmental Program in Biomolecular Science and
Engineering and 6Department of Chemistry and Biochemistry, University of
California, Santa Barbara, CA 93106; Departments of 2Applied Physics and
5Chemistry, Stanford University, Stanford, CA 92343; 3Department of
Biochemistry and Molecular Biology and 8Institute for Biophysical Dynamics,
University of Chicago, Chicago, IL 60637; 4Intense Pulsed Neutron Source and
7Advanced Photon Source, Argonne National Laboratory, Argonne, IL 60439;
9Department of Physics, Princeton University, Princeton, NJ 08544; and
10Department of Biostatistics, Bloomberg School of Public Health, The Johns
Hopkins University, Baltimore, MD 21205
This research was supported by the National Institutes of Health (K.W.P. and
V.S.P.), the Packard Foundation Interdisciplinary Science Program (T.R.S.
and P.T.), and a faculty innovation award from The Johns Hopkins University
(to I.R.).